Loop calling¶
Loops frequently form between two genomic regions, and are visible in the Hi-C matrix as patches of increased contact intensity:
fancplot -o architecture/loops/rao2014.chr11_77400000_78600000.png \
chr11:77400000-78600000 \
-p triangular architecture/loops/rao2014.chr11_77400000_78600000.hic \
-vmin 0.0 -vmax 0.05 -m 600000
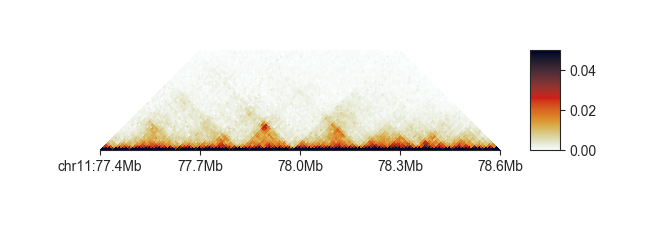
We can use fanc loops to call loops in Hi-C matrices using the HICCUPS algorithm
(Rao and Huntley et al., 2014).
Please refer to the original paper for details on the algorithm, specifically the
different types of local neighborhoods defined to make loop calling robust.
usage: fanc loops [-h] [-c CHROMOSOMES [CHROMOSOMES ...]] [-p PEAK_SIZE]
[-w WIDTH] [-t THREADS] [-d MIN_DIST]
[-m MAPPABILITY_GLOBAL_CUTOFF]
[--mappability-donut MAPPABILITY_DONUT_CUTOFF]
[--mappability-horizontal MAPPABILITY_HORIZONTAL_CUTOFF]
[--mappability-vertical MAPPABILITY_VERTICAL_CUTOFF]
[--mappability-lower-left MAPPABILITY_LOWER_LEFT_CUTOFF]
[-q FDR_GLOBAL_CUTOFF] [--fdr-donut FDR_DONUT_CUTOFF]
[--fdr-horizontal FDR_HORIZONTAL_CUTOFF]
[--fdr-vertical FDR_VERTICAL_CUTOFF]
[--fdr-lower-left FDR_LOWER_LEFT_CUTOFF]
[-e ENRICHMENT_GLOBAL_CUTOFF]
[--enrichment-donut ENRICHMENT_DONUT_CUTOFF]
[--enrichment-horizontal ENRICHMENT_HORIZONTAL_CUTOFF]
[--enrichment-vertical ENRICHMENT_VERTICAL_CUTOFF]
[--enrichment-lower-left ENRICHMENT_LOWER_LEFT_CUTOFF]
[-o OBSERVED] [--rh-filter] [--sge]
[--batch-size BATCH_SIZE] [-j]
[--merge-distance MERGE_DISTANCE] [-s] [--fdr-sum FDR_SUM]
[-b BEDPE] [-f] [-tmp]
input [output]
Positional Arguments¶
- input
Input Hic file
- output
Output file. If input file is already a FAN-C compatible loops object, filtering can also be done in place.
Named Arguments¶
- -c, --chromosomes
Chromosomes to be investigated.
- -p, --peak-size
Size of the expected peak in pixels. If not set, will be estimated to correspond to ~ 25kb.
- -w, --neighborhood-width
Width of the investigated area surrounding pixels. If not set, will be estimated at p+3
- -t, --threads
Number of threads for parallel processing. Default: 1 - it is advised to set this as high as possible, since loop calling is very computationally expensive!
- -d, --min-distance
Minimum distance in bins for two loci to be considered as loops. Default: peak size. Set this value higher to exclude loops close to the diagonal.
- -m, --mappability
Global mappability filter for all neighborhoods.Minimum mappable fraction of a pixel neighborhood to consider pixel as loop. Can be overridden by filters for local neighborhoods.
- --mappability-donut
Mappability filter for donut neighborhood. Value between 0 and 1.
- --mappability-horizontal
Mappability filter for horizontal neighborhood. Value between 0 and 1.
- --mappability-vertical
Mappability filter for vertical neighborhood. Value between 0 and 1.
- --mappability-lower-left
Mappability filter for lower-left neighborhood. Value between 0 and 1.
- -q, --fdr
Global FDR filter all neighborhoods. Individual neighborhood filters can override this global setting. Value between 0 and 1.
- --fdr-donut
FDR filter for donut neighborhood. Value between 0 and 1.
- --fdr-horizontal
FDR filter for horizontal neighborhood. Value between 0 and 1.
- --fdr-vertical
FDR filter for vertical neighborhood. Value between 0 and 1.
- --fdr-lower-left
FDR filter for lower-left neighborhood. Value between 0 and 1.
- -e, --enrichment
Global observed/expected filter all neighborhoods. Individual neighborhood filters can override this global setting.
- --enrichment-donut
Observed/expected enrichment filter for donut neighborhood.
- --enrichment-horizontal
Observed/expected enrichment filter for horizontal neighborhood.
- --enrichment-vertical
Observed/expected enrichment filter for vertical neighborhood.
- --enrichment-lower-left
Observed/expected enrichment filter for lower-left neighborhood.
- -o, --observed
Minimum observed value (integer, uncorrected). Default: 1
- --rh-filter
Filter peaks as in Rao, Huntley et al. (2014), Cell. It only retains peaks that are at least 2-fold enriched over either the donut or lower-left neighborhood, at least 1.5-fold enriched over the horizontal and vertical neighborhoods, at least 1.75-fold enriched over both the donut and lower-left neighborhood, and have an FDR <= 0.1 in every neighborhood
- --sge
Run on SGE cluster. This option is highly recommended if you are running “fanc loops” on a Sun/Oracle Grid Engine Cluster. The “-t” option specifies the number of SGE slots if this flag is set. The main process will then submit jobs to the grid using “gridmap” and collect the results. (https://gridmap.readthedocs.io/)
- --batch-size
Width of submatrix examined per process. Default: 200
- -j, --merge-pixels
Merge individual pixels into peaks after filtering.
- --merge-distance
Maximum distance in base pairs at which to merge two pixels. Default 20000
- -s, --remove-singlets
Remove isolated pixels after merging step.
- --fdr-sum
FDR sum filter for merged peaks. Merged peaks where the sum of donut FDR values of all constituent pixels is larger than the specified cutoff are filtered.
- -b, --bedpe
BEDPE output file. When set, merged loops will be written to this file after all filtering steps have completed.
- -f, --force-overwrite
- If the specified output file exists, it will be
overwritten without warning.
- -tmp, --work-in-tmp
Work in temporary directory
Annotating pixels for loop calling¶
The first step in HICCUPS consists of annotating each pixel with various measures related to their loop probability. The most important ones are
Enrichment over expected values in the local neighborhood
FDR of the local enrichment
Mappability of the local neighborhood
fanc loops architecture/loops/rao2014.chr11_77400000_78600000.hic \
architecture/loops/rao2014.chr11_77400000_78600000.loops \
-t 2
When run like this, fanc loops does not actually call any loops, but merely returns
a matrix object where every pixel is annotated with the above properties. Importantly,
as this is the most computationally expensive step, you are strongly advised to choose
a large number of threads using the -t option. Even better, if you have access to
a computational cluster running Sun/Oracle Grid Engine, fanc loops can automatically
submit annotation jobs to the cluster if you set the --sge flag. The -t option
then specifies the number of jobs allowed to run in parallel instead of the number of
local threads used for multiprocessing.
By default, fanc loops assumes a loop size of 25kb. This determines the area around
a pixel that is not included in the local neighborhood calculations. If this is chosen
too small, the neighborhood will lie within the peak region, and enrichments are going
to be lower. If this is chosen too big, the neighborhood will no longer be local. If you
have reason to believe your loops size differes from the default, you can set it explicitly
with -p.
Similarly, the width of the neighborhood is determined as p + 3 by default. If you want
to in- or decrease the neighborhood width, use the -w parameter. You should know,
however, that this is just a starting value, and the neighbborhood width might be increased
on demand internally.
Finally, you can control the size of the submatrices sent to each thread using the
--batch-size parameter. The default, 200, should suit most purposes, but if your
individual jobs are taking too long, you should reduce this number.
We can now use the output object with annotated pixels for downstream processing.
Filtering annotated pixels¶
We need to apply filters to the annotated pixel object that remove all pixels with a low probability of being a loop. These filters typically consist of enrichment filters, FDR filters, and mappability filters. Additionally, there are filters for minimum distance between regions, and the minimum number of unnormalised valid pairs in a pixel.
You can either set a global enrichment filter that acts on all neighborhoods
using -e, or choose individual thresholds for each local neighborhood with
--enrichment-donut, --enrichment-vertical, --enrichment-horizontal,
and --enrichment-lower-left. You usually want to set at least the
--enrichment-donut cutoff to something like 2.
For FDR values, also called q-values, you can set a global filter using -q. Control
the filtering of individual neighborhoods using --fdr-donut, --fdr-vertical,
--fdr-horizontal, and --fdr-lower-left. Typical values for each neighborhood
are around 0.1.
Mappability filters act on pixels where a certain fraction of pixels in their local
neighborhoods is unmappable. To set a global mappability cutoff for all neighborhoods,
use the -m option. Again, local neighborhood mappability filters can be fine-tuned
using the --mappability-donut, --mappability-vertical, --mappability-horizontal,
and --mappability-lower-left options.
It is generally a good idea to filter on the minimum distance between regions to
consider forming a loop, as a lot of false positive loops will be close to the diagonal.
You can use the -d <b> parameter to set a threshold on the minimum distance, where
b is expressed in number of bins.
In addition, we highly recommend applying a filter on the minimum number of valid pairs
in a pixel (-o), so that false-positive loops due to noise are avoided.
Finally, we have included the filter applied by Rao and Huntley et al. in their original
HICCUPS algorithm as a convenient preset --rh-filter. It only retains peaks that are
at least 2-fold enriched over either the donut or lower-left neighborhood, at least
1.5-fold enriched over the horizontal and vertical neighborhoods, at least 1.75-fold
enriched over both the donut and lower-left neighborhood, and have an FDR <= 0.1 in
every neighborhood.
An example command could look like this:
fanc loops architecture/loops/rao2014.chr11_77400000_78600000.loops \
architecture/loops/rao2014.chr11_77400000_78600000_filtered.loops \
--rh-filter -d 5 -o 5
This filters the vast majority of pixels in the matrix.
Merging unfiltered pixels into loops¶
Pixels that pass all filtering steps are good candidates for loops. Often, these pixels
appear in clusters, which we merge/join in this step. Pixels that do not form a cluster
are generally false-positives, so we filter them using --remove-singlets.
fanc loops architecture/loops/rao2014.chr11_77400000_78600000_filtered.loops \
architecture/loops/rao2014.chr11_77400000_78600000_merged.loops \
-j --remove-singlets
Exporting to BEDPE¶
Finally, we can export all the merged loops to BEDPE using -b:
fanc loops architecture/loops/rao2014.chr11_77400000_78600000_merged.loops \
-b architecture/loops/rao2014.chr11_77400000_78600000_merged.bedpe